Title of the project: Knowledge-Aware Data Augmentation for Transcriptomics
Laboratory:
IBISC (Informatique, Bioinformatique et Systèmes Complexes), Université d’Évry, Université de Paris-Saclay
Project coordinator and thesis supevisor:
Blaise HANCZAR, Professeur des universités, Université d’Évry, Université de Paris-Saclay, IBISC, AROB@S team.
Thesis co-supervisor
Michèle SEBAG, Directrice de Recherche CNRS, Université de Paris-Saclay, LISN A&O team.
Context
This proposal considers domains where only small data are available, and it specifically focuses on transcriptomics. Transcriptomic data represents the amount of RNA « produced » by each gene, called gene expression; it can be viewed as the activity of genes in a sample. These data are the basis of numerous studies and biomedical applications such as the analysis of regulatory networks, the identification of the biological role of genes, the aid in the diagnosis/prognosis of a patient, the estimation of the effect of a treatment [1]. Analysis of transcriptomic data plays a major role in understanding molecular biology and the development of personalized medicine. Due to the high cost of data production however, transcriptomic datasets contain few samples: the size of the datasets ranges from a few hundred to a thousand.
While machine learning methods are commonly used to exploit transcriptomic data in a satisfactory way, e.g. for phenotype prediction tasks [2], the obtained results lack robustness and they are found difficult to reproduce; these difficulties are blamed on the small dataset size. Most generally, the shortage of data prevents the deployment of approaches designed for Big Data, e.g., deep learning.
State of the art
Several research directions have been investigated to address such a shortage of data. In the case where some rich source dataset, “sufficiently similar” to the considered target dataset, is available, domain adaptation [3,4] leverages the source data to learn more robust models on the target data. In multi-task learning [5], heterogeneous datasets, possibly gathered along different distributions and involving different classes, are reconciled and mapped onto a shared representation, supporting the learning of general and robust models.
Another approach, data augmentation uses domain knowledge and/or heuristics to generate additional data. For instance, including new images obtained by vertical symmetry from the original ones, is a simple way to increase the dataset size, and increase the predictive accuracy of the learned models, e.g. when identifying rare birds or insects. Data augmentation can thus be viewed as a regularization in extension, exploiting in this case the fact that the category of bird or insect is known to be invariant under vertical symmetry of the image.
Objectives
The objective of the KADAT project is to design a principled data augmentation process, taking advantage of the fact that small data domains often enjoy rich domain knowledge. This knowledge-aware data augmentation is intended to yield more robust models from small datasets, through making these models compliant with the domain knowledge. The originality is that this compliance is enforced in extension and a priori (through the exploitation of additional examples) as opposed to, in intention and a posteriori (editing the models to comply with the knowledge).
Specifically in the context of transcriptomic data, an extensive and regularly updated knowledge base, the Gene Ontology (GO) is available and represents current knowledge on the role of genes. Formally, GO is a directed acyclic graph containing all the biological functions, molecular processes and cellular components. Each node corresponds to one of these entities with the genes for which a link has been identified, the edges correspond to relations between to biological entities.
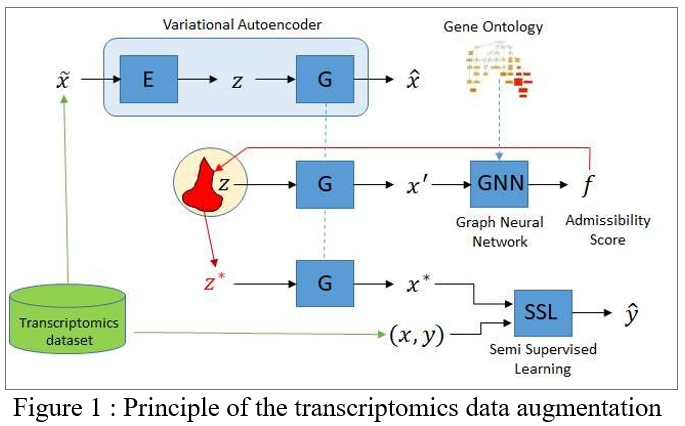
The Knowledge-Aware Data Augmentation for Transcriptomics (KADAT) project will proceed as follows. The first milestone consists in defining an estimate of the admissibility of a true data sample, learned from the initial data. A first challenge consists of learning such an estimate from the initial dataset (of limited size by construction). This challenge is tackled, taking advantage of the modularity of the knowledge base (see below); intuitively, the idea is that each knowledge “nugget” is associated with a local admissibility score, trained from the initial dataset.
The second milestone consists in biasing a Variational Auto-Encoder (VAE) [6], to enforce the generation of new data yielding satisfactory admissibility scores. The second challenge is to train a VAE from the initial dataset, granted that deep learning and VAEs notoriously require large data resources. This challenge can be addressed using a naive data augmentation, e.g. based on the addition of small Gaussian noise to each instance in the dataset. The VAE trained from this initial augmentation is thereafter biased to yield new samples with a high admissibility score. Note that these new samples do not come with a label (see below).
The augmented dataset will be exploited along a semi-supervised setting [7], considering the true examples (with their labels) and the augmented examples (with no labels). The regularization classically proceeds by requiring that the sought classifier yields a sufficient margin on the augmented examples (the frontier induced by the classifier is required to be located in low density regions). This regularization, akin an additional constraint on the classifier, is expected to result in more robust and stable classifiers.
If time permits, the potential of this original data augmentation process, that is original to our best knowledge, will be investigated in the context of domain adaptation and multi-task learning algorithms; the expectations likewise are that the data augmentation will contribute to the robustness of the results.
The merits of the approach will be measured by comparing the performance accuracy of the classifiers learned from the datasets augmented with KADAT and with a naive augmentation process (e.g. based on the addition of Gaussian noise).
Methods
Milestone 1: Learning an admissibility score.
The envisioned approach builds upon Graph Neural Nets (GNN) [8]. The architecture of this GNN will emulate the DAG structure of the GO ontology, where each node corresponds to a biological entity and an edge among nodes reflects the presence of a relationship between both entities. Each data sample (d-dimensional real vector) describes the level of activity of some of these nodes. Based on these data samples, the parameters of the activation function in each node will be trained, describing how the activity in a node can be predicted from the activity of its neighbor nodes. Note that GO nodes are not necessarily all represented in the dataset: these will be handled as latent variables. Given the limited number of edges per node (circa 10) in GO, it is believed that the parameters of each activation function can be learned in a reasonably robust and efficient manner from a few hundred sample dataset.
The trained GNN can be used to infer an admissibility score on any d-dimensional vector, for instance by considering its mean square prediction error (squared difference between the activity predicted for a node, depending on the actual activity of its neighbor nodes, and its actual activity), akin an auto-encoder.
Milestone 2: Biasing a Variational Auto-Encoder.
The standard VAE framework is considered. Denoting p the distribution defined on the instance space (the initial VAE) and f the admissibility score, to be maximized, one aims to find a new distribution q such that it maximizes the expectation of f subject to remaining close to p in terms of Kullback-Leibler divergence:
This above constrained optimization problem will be solved, using a Lagrangian formulation (see also [9]). Most interestingly, this optimization problem can also be solved in the latent space of the VAE, thus with a reduced dimensionality compared to that of the instance space.
Datasets
We consider two transcriptomic datasets. The first one is the result of a cross-experimental study compiling circa 20,000 microarrays from DNA chips under different experimental conditions, where the objective is to predict the presence of cancer in a tissue [10]. The second one comes from the TCGA program combining the RNA-seq gene expression of circa 10.000 patients of 33 cancer types [11]. The objective is to predict the type of cancer or the prognostic of a patient. The comparatively large size of these datasets will allow a rigorous experimental validation of the approach, based on training sets aggressively subsampling the overall dataset.
Team
This project involves the AROBAS (Algorithmique, Recherche opérationnelle, Bioinformatics, Apprentissage Statistique) team from IBISC, Université d’Evry Université Paris-Saclay and the A&O (Apprentissage et Optimisation) team from LISN, Université Paris-Saclay.
Blaise Hanczar, who is a specialist in machine learning for genomics data, will be the main PhD supervisor and lead the work on graph neural networks; Michele Sebag, who is expert in deep learning, will lead the generative model part. The partners of this project have successfully collaborated 6 years ago [12].
Profil of the PhD Student
The student will have a Master in Machine Learning (in Computer Science or Applied Mathematics). An interest or prior experience in bioinformatics will be appreciated.
Bibliography
[1] Xu, J., Yang, P., Xue, S., Sharma, B., Sanchez-Martin, M., Wang, F., … & Parikh, B. (2019). Translating cancer genomics into precision medicine with artificial intelligence: applications, challenges and future perspectives. Human Genetics, 138(2), 109-124.
[2] Kourou, K., et al. (2015). Machine learning applications in cancer prognosis and prediction. Computational and structural biotechnology journal, 13, 8-17
[3] Ben-David, S., Blitzer, J., Crammer, K., & Pereira, F. (2007). Analysis of representations for domain adaptation. In NIPS, (pp. 137-144).
[4] Ganin, Y., et al., (2016). Domain-Adversarial Training of Neural Networks. Mach. Learn. Res. 17: 59:1-59:35.
[5] Schoenauer-Sebag, A., et al. (2019). Multi-domain adversarial learning. ICLR.
[6] Kingma, D. P., & Welling, M. (2019). An introduction to variational autoencoders. arXiv preprint arXiv:1906.02691.
[7] Miyato, T., Maeda, S. I., Koyama, M., & Ishii, S. (2018). Virtual adversarial training: a regularization method for supervised and semi-supervised learning. IEEE transactions on pattern analysis and machine intelligence, 41(8), 19791993
[8] Wu, Z., Pan, S., Chen, F., Long, G., Zhang, C., & Philip, S. Y. (2020). A comprehensive survey on graph neural networks. IEEE Transactions on Neural Networks and Learning Systems.
[9] Hu, Z. et al., (2018). Deep Generative Models with Learnable Knowledge Constraints, NeurIPS 18.
[10] Torrente, A., et al. (2016). Identification of cancer related genes using a comprehensive map of human gene expression. PloS one, 11(6), e0157484.
[11] https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
[12] Hanczar, B., & Sebag, M. (2014). Combination of one-class support vector machines for classification with reject option. In ECML-PKDD, Springer, (pp. 547-562).
- Date de l’appel : 05/05/2021
- Statut de l’appel : Non pourvu
- Contact : Blaise HANCZAR (PR Univ. Évry, IBISC équipe AROB@S)
- Sujet de thèse (format PDF)
- Web équipe AROB@S




