For my benchmarks, I used to store logs of every executions into CSV files that I had to aggregate for analysis. I’ve decided to switch to a database. It is simpler and globally more efficient.
Discussion
My benchmarks are usually organised as follows:
- there are several models
- each one may be instantiated with a distinct set of parameters, each such instance is a case to be benchmarked
- each case is implemented as a Petri net (so surprising)
- the goal is to compare different tools on all the cases
- each Petri net is built in various ways depending on the tool that will use it
- for each tool and each case, the corresponding net is used to exercise the tool
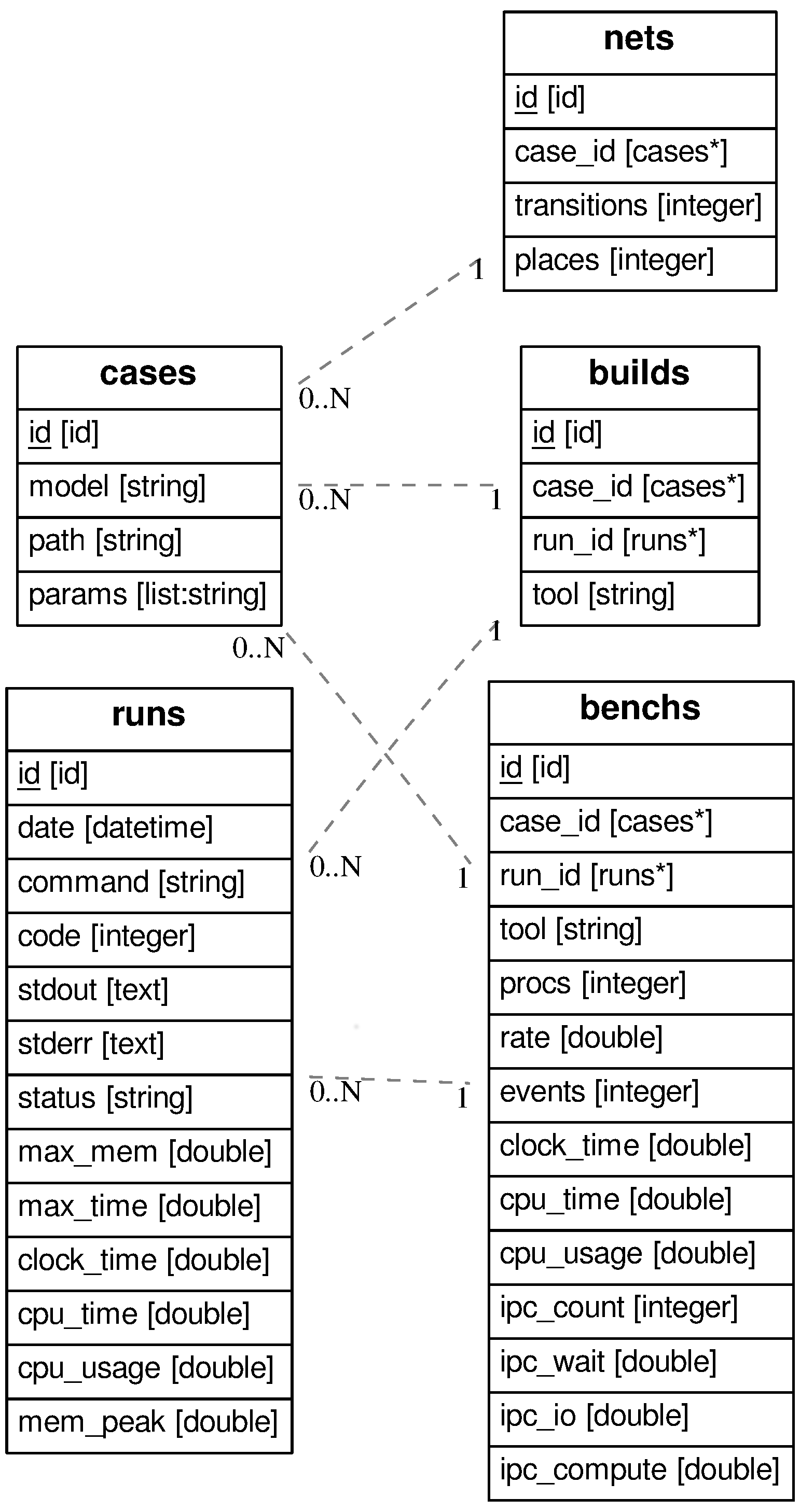 I’m not a database enthusiast, actually, I must confess I don’t like
databases. But to be honest, this situation is typically the kind of
problems databases have been made to solve. So, let’s go. The
relational diagram is shown here on the right, (drawn from a
particular example I’m working on
using ERAlchemy). We
see that we have tables corresponding to the objects described above,
plus a table
I’m not a database enthusiast, actually, I must confess I don’t like
databases. But to be honest, this situation is typically the kind of
problems databases have been made to solve. So, let’s go. The
relational diagram is shown here on the right, (drawn from a
particular example I’m working on
using ERAlchemy). We
see that we have tables corresponding to the objects described above,
plus a table runs that records the information related to any tool
execution. I won’t go into the details since it doesn’t serve my
topic. How this is implemented is described below, with code excerpts,
but note that parts of the remarks below is dependent on this
implementation.
So, what have we gained now we have a database?
First, a database is a storage system much simpler to manage that tons
of CSV files, in particular, we don’t have to manage how the different
data types are encoded. I can forget about the column types, even
complex ones like list:string that is automatically handled.
Moreover, the definition of the database includes integrity
constraints, for example a net cannot refer to a case that does not
exist.
Then, a database can be queried. Here, I can easily get all the cases for which a net exists. Or: all the cases for a given model for which there is a net with at most n transitions that could be built within at most 10 seconds, ordered by the number of transitions. With CSV files, I would have to load all the data, compile it into a single Pandas table (for instance) and then perform the query on the table, which is possible of course, but not as convenient as with the database (and probably less efficient).
Moreover, with CSV files, I have to compute the joins myself, either while dumping the data (for instance, having one series of files that joins cases, nets, builds, and runs, plus another series for cases, nets, benchs, and runs), or by computing the joins after loading the data. The first solution is simpler but wastes disk space, especially for thousands or runs; the second solution is complicated and requires delicate programming. Of course Pandas can help a lot to compute joins, but again, it’s not its job.
Finally, the storage is more efficient in a database and, using SQLite in particular, it’s easier to copy around various versions of the database. For example, I copy it from the cluster that runs the benchmark to my laptop on which I do the data analysis.
Conclusion
There is no definitive argument in favour of using a database, but my experience is that it’s a better tool because its designed exactly for this purpose. And it’s not more complicated using the right libraries as shown below.
Implementation
Database
I’ve used pyDAL that is the standalone version of the database abstraction layer from the excellent web2py. Here is the code to declare the model depicted above:
from pydal import DAL, Field
def opendb (path="benchmark.db") :
db = DAL("sqlite://" + path)
db.define_table("cases",
Field("model", "string"),
Field("path", "string", unique=True),
Field("params", "list:string"))
db.define_table("nets",
Field("case_id", "reference cases"),
Field("transitions", "integer"),
Field("places", "integer"))
db.define_table("runs",
Field("date", "datetime"),
Field("command", "string"),
Field("code", "integer"),
Field("stdout", "text"),
Field("stderr", "text"),
Field("status", "string"),
Field("max_mem", "double"),
Field("max_time", "double"),
Field("clock_time", "double"),
Field("cpu_time", "double"),
Field("cpu_usage", "double"),
Field("mem_peak", "double"))
db.define_table("builds",
Field("case_id", "reference cases"),
Field("run_id", "reference runs"),
Field("tool", "string"))
db.define_table("benchs",
Field("case_id", "reference cases"),
Field("run_id", "reference runs"),
Field("tool", "string"),
Field("procs", "integer"),
Field("rate", "double"),
Field("events", "integer"),
Field("clock_time", "double"),
Field("cpu_time", "double"),
Field("cpu_usage", "double"),
Field("ipc_count", "integer"),
Field("ipc_wait", "double"),
Field("ipc_io", "double"),
Field("ipc_compute", "double"))
return db
So this is really straightforward. I’ve chosen SQLite backend so that there is no database installation at all and it is easy to copy it between the cluster and my laptop.
One very interesting feature of pyDAL is that it does automatic migration of an existing database. So if I decide to add or remove tables or columns, it will convert the existing data into the new structure. (There’s no magic, if you completely transform the structure, it won’t work automatically.)
Now, this is the code to convert the result of any request into a
Pandas DataFrame.
import pandas as pd
def row2dict (row) :
"Convert a row into a flat dict"
ret = {}
for key, val in row.as_dict().items() :
if key == "id" or key.endswith("id") or val is None :
# don't copy database ids
pass
elif isinstance(val, list) :
# parameters in lists are encoded as key=value
for v in val :
k, d = v.split("=", 1)
try :
d = ast.literal_eval(d)
except :
pass
ret[k] = d
elif isinstance(val, dict) :
# recursively decode a row
ret.update(row2dict(row[key]))
elif isinstance(val, (str, int, long, float, datetime.datetime)) :
# simple values
ret[key] = val
else :
# could be extended here
assert False, "unsupported %s=%r" % (key, val)
return ret
def db2pd (rows) :
"Convert a list of rows into a DataFrame"
return pd.DataFrame(row2dict(r) for r in rows)
So, for instance, to analyse how the nets have been generated, I can do:
df = db2pd(db((db.cases.id == db.nets.case_id)
& (db.cases.id == db.builds.case_id)
& (db.runs.id == db.builds.run_id)).select())
Ordering the results by transitions just requires to add
orderby=db.nets.transitions as a select() argument. More
generally, pyDAL offers a rich set of possibilities to query the
databases, and it is of course possible to evaluate raw SQL as well.
Measuring process performances
This is not the topic of the post, but here is the code I use to run a process while measuring its CPU and memory usage.
import collections, psutil, subprocess, datetime, os, time
# information about a process run
run = collections.namedtuple("run", ["date", "command", "status", "code",
"clock_time", "cpu_time", "cpu_usage",
"mem_peak", "stdout", "stderr"])
def newrun (oldrun, **update) :
"Build a new run updating some fields"
return run(*(update.get(attr, oldrun[pos])
for pos, attr in enumerate(oldrun._fields)))
def spawn (*command, timeout=0, maxmem=90, env=None, cwd=None) :
"Run command and return the corresponding run object"
command = [str(c) for c in command]
if env is not None :
newenv = os.environ.copy()
newenv.update(env)
env = newenv
status = None
peak = 0
main = psutil.Popen(["time", "-f", "%%%e/%S/%U/%P"] + command,
stdout=subprocess.PIPE, stderr=subprocess.PIPE,
cwd=cwd, env=env)
time.sleep(0.01) # let the process start
child = main.children()[0]
start = main.create_time()
while True :
try :
# memory usage is measured every 1 second, this may be
# sampled more often for short living processes
if main.wait(1) is not None :
break
except psutil.TimeoutExpired :
# process has not finished
pass
try :
# get memory usage
peak = max(peak, child.memory_info().vms)
pmem = child.memory_percent()
except psutil.NoSuchProcess :
# process has finished
break
if timeout and time.time() - start > timeout :
# process runs out of time
status = "reached time limit"
elif maxmem and pmem > maxmem :
# process consumes too much memory (90% by default)
status = "reached mem limit"
if status :
# kill the process because of timeout/maxmem
try :
# SIGTERM (be gentle)
child.terminate()
except :
pass
try :
# give it 1 second to stop
child.wait(1)
except :
pass
try :
# SIGKILL (enough is enough)
child.kill()
except :
pass
# parse stderr to get the output from time utility
stderr, times, _ = main.stderr.read().rsplit("%", 2)
times = times.split("/")
if times[3] == "?" :
times[3] = "0"
return run(date = datetime.datetime.fromtimestamp(start),
command = command,
status = status or "clean",
code = main.returncode,
clock_time = float(times[0]),
cpu_time = float(times[1]) + float(times[2]),
cpu_usage = float(times[3]) / 100.0,
mem_peak = peak,
stdout = main.stdout.read(),
stderr = stderr)
def pyspawn (*command, **options) :
"Same as spawn but also parse Python exception traceback"
t = spawn(*command, **options)
status = None
if t.code != 0 and t.status == "clean" :
if any(l == "MemoryError" for l in t.stderr.splitlines()) :
status = "out of memory"
elif any(l == "Traceback (most recent call last):"
for l in t.stderr.splitlines()) :
status = "raised Exception"
lines = iter(t.stderr.splitlines())
for line in lines :
if line == "Traceback (most recent call last):" :
break
for line in lines :
if not line.startswith(" ") :
status = "raised " + line
break
else :
status = "error"
return run(date = t.date,
command = t.command,
status = status or t.status,
code = t.code,
clock_time = t.clock_time,
cpu_time = t.cpu_time,
cpu_usage = t.cpu_usage,
mem_peak = t.mem_peak,
stdout = t.stdout,
stderr = t.stderr)
def run2dict (run) :
"""Convert a run into a dict for easy insertion in the database
For example:
>>> r = run(...)
>>> db.runs.insert(**run2dict(r))
"""
return dict(zip(run._fields, run))
Drawing the database schema
Last, just to show how ERAlchemy is used:
def db2diagram (db, path) :
"Draw the relational diagram of a database"
with open(path, "w") as out :
links = {}
for table in db.tables :
out.write("[%s]\n" % table)
for field in db[table].fields :
if field == "id" :
star = "*"
else :
star = ""
kind = getattr(db[table], field).type
if kind.startswith("reference ") :
_, target = kind.split()
links[target, table] = "*--1"
kind = "%s*" % target
out.write(' %s%s {label: "%s"}\n' % (star, field, kind))
for (src, tgt), lnk in links.items() :
out.write("%s %s %s\n" % (src, lnk, tgt))
os.system("eralchemy -i %s -o %s.pdf" % (path, os.path.splitext(path)[0]))