Sujet : « Offre de stage Master 2, équipe SIAM : « Medical imaging for the prediction of survival of patients with stroke »
Keywords
machine learning, deep tech, neuroimaging, precision medicine, stroke
Context and objectives
According to the World Health Organization, stroke is the second cause of death and the leading cause of chronic functional disability in adults, with 17 million victims, 31% of whom were under the age of 65. More than 6 million people die from stroke worldwide each year.
In France, each year around 150,000 people are hospitalized for a stroke, one every 4 minutes with an average cost of 19 k€. It is estimated that 750,000 people have survived a stroke, two thirds of which will have disabling consequences, which represents a financial burden for the state of around 2.8 billion €/year… in reality 10 billion over 5 years due to the cost of handicap.
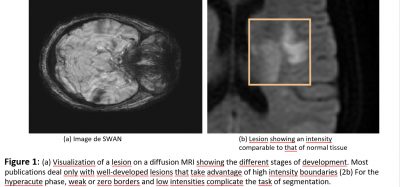
The ischemic stroke is caused by a blood clot (thrombus) that blocks a brain artery causing lack of oxygen brain tissue supplied by that artery (Fig. 1). There is an urgent need to diagnose and determine if treatment with thrombolytic drugs (anti-coagulants) can “reverse” the stroke. The response time is limited and should not exceed 3 to 4 hours after the onset of symptoms. Confronted with the management of a stroke, the doctor then asks 3 questions to which the imagery provides particularly relevant answers: is it really a stroke? Is the stroke ischemic or hemorrhagic in nature? If thrombolysis is considered, are there any radiological contraindications to this treatment? There is consensus that magnetic resonance imaging (MRI) is the gold standard for eliminating non-vascular diagnoses because of its sensitivity and specificity in acute ischemia.
Hospital reception therefore favors the speed of access to the neurovascular unit (NVU) and MRI to confirm the diagnosis of cerebral infarction or cerebral hemorrhage: early treatment (<3h) limits the severity sequelae. If MRI makes it possible to search for the cause of the lesion, it raises many methodological difficulties linked to the very progressive pathophysiology of stroke in the very first hours:
- a sequence of complex multifactorial images from which the doctor must decide on a treatment in a few minutes
- the operator character depending on the technique considered.
- the reduced definition of images due to the urgency of the diagnosis. The early diagnosis of stroke by MRI represents a big data challenge. An automatic method to segment the lesion would improve MRI interpretation and productivity, by quantifying the relevant parameters instead of the current approximations. However, there has not been a complete automatic tool for the simultaneous segmentation of lesions to date.
Objectives
The solution that we want to implement is based on the automatic segmentation of irreversible infarction areas and ischemic tissues at risk, but also on the localization of the thrombus at the origin of these lesions thanks to deep learning algorithms from artificial intelligence. The application of these algorithms to the analysis of solid images makes it possible to work on a large amount of data in a more relevant way than conventional statistical methods. The objectives are to validate the results on a large patient database and to integrate the model into clinical application software with a user-friendly interface.
Methodology No method of automatic segmentation of the lesion has been published to date. There are a few publications on semi-automatic segmentation of the lesion by angiography [4, 10, 7, 12, 11]. Only the last three relate to the segmentation of the lesion in the brain. What they all have in common is that they need a “manual” seed to run the algorithm, that is, they cannot find the position of the lesion on their own.
Qazi et al. [10] study segmentation via thresholds, but this is not adaptable to MRI because the lesion in a SWAN is not segmentable by a simple threshold. Among current segmentation algorithms, the CNN has been much tried for the segmentation of lesions. For example, almost all of the ISLES 2017 brain injury segmentation challenge submissions were based on CNN with variable architecture, and with rather poor results (the best reported DICE was 0.31 [3]). An example of an efficient architecture for assigning a probability of injury to each voxel was proposed by Chen et al. [2] with good segmentation results over a wide range of lesion sizes and intensity variations.
But their algorithm cannot separate lesions and artifacts if they are connected to the image. And they missed some of the very weak parts of the lesion.
The reason is the lack of large MRI ischemic data sets available due to their cost of acquisition and anotation. The difficulty of obtaining a sufficient amount of reliable class-specific training data for a supervised automatic approach requires the study of new strategies. A solution suggested by very recent studies [3], proposes to develop new generic salience functions or to use the data augmentation method to build a robust classification as well as other parameters such as texture or shape. Learning by neural networks inspired by ladder networks or regime networks or adversary autoencoding, curicular model, etc. will be privileged in this internship.
Expected results
The expected solution will better characterize stroke by associating multiple weak signals with the definition of pathology, which can therefore lead to replacing complex clinical scales (CPSSS, NIHSS, LAMS, VAN, etc.), which are tedious to calculated in current practice. It will improve the quality of reading of current images, for example, by performing analyzes which are not currently carried out because they take too long to execute manually such as the volumetric measurement of the lesion, the extraction of textures, etc. This solution will be able to determine what information in the image implies certain treatments leading to better results for patients. AI can help the radiologist prioritize urgent cases by determining in advance which imaging tests to assess first. AI can do an initial assessment and escalate cases if necessary. Ultimately, the technique should give more information than the human eye on the texture of the thrombus and its accessibility for recanalization treatments.
Expected performance criteria:
Evaluating the new procedure against a referenced procedure raises many methodological difficulties. The expected performance indicators are
- the repeatability of the (deterministic) segmentation process in a degraded situation or not,
- the efficiency of the tool to be tested on a ground truth basis and quantified with DICE [3] to measure performance in segmentation, 3. a speed of execution of a few minutes.
Profile and skills required
Ability to understand and develop adaptive learning algorithms and to process medical data, index it and use it in an operational system to achieve the mission described above.
Programming skills: Python or C / C ++. A practice of Tensorflow and Pytorch would be a plus. The practice of French is not compulsory. His(her) English is fluent. The work will be carried out at the IBISC Laboratory located on the Evry campus of the UPSaclay. IBISC develops multidisciplinary, theoretical and applied research in the field of information sciences and engineering, with a strong orientation towards health applications. The selected candidate will have the chance to work in an interdisciplinary team and with a consortium of data scientists and clinicians from the CHSF. The project is multidisciplinary, at the interface of machine learning, computer science and medicine.
Encadrement et conditions scientifiques et matérielles
The student will be supervised by Vincent Vigneron, Jean-Philippe Congé and Hichem Maaref from the BISC laboratory (Univ d’Évry, Université Paris-Saclay). All master machine learning, signal and image processing.
Vincent Vigneron and Hichem Maaref {hichem.maaref,vincent.vigneron}@univevry.fr
Phone: +33 6 635 687 60
date de début et de fin du stage 15/02/2022 au 1/09/2022
References
- [1] Ikram Brahim, Dominique Fourer, Vincent Vigneron, and Hichem Maaref. Deep Learning Methods for MRI Brain Tumor Segmentation: a comparative study. In 9th IEEE International Conference on Image Processing Theory, Tools and Applications (IPTA 2019), Istanbul, Turkey, November 2019.
- [2] Liang Chen, Paul Bentley, and Daniel Rueckert. Fully automatic acute ischemic lesion segmentation in dwi using convolutional neural networks. NeuroImage: Clinical, 15:633 – 643, 2017.
- [3] Lee R. Dice. Measures of the amount of ecologic association between species. Ecology, 26(3):297–302, 1945.
- [4] Egger, Thomas O’Donnell, C. Hopfgartner, and B. Freisleben. Graph-based tracking method for aortic thrombus segmentation. In Jos Vander Sloten, Pascal Verdonck, Marc Nyssen, and Jens Haueisen, editors, 4th European Conference of the International Federation for Medical and Biological Engineering, pages 584–587, Berlin, Heidelberg, 2009. Springer Berlin Heidelberg.
- [5] Jonathan Kobold, Vincent Vigneron, Hichem Maaref, Dominique Fourer, M Aghasaryan, C Alecu, N Chausson, Y L’hermitte, D. Smadja, and E. Läng. Stroke Thrombus Segmentation on SWAN with MultiDirectional U-Nets. In 9th IEEE International Conference on Image Processing Theory, Tools and Applications (IPTA 2019), Istanbul, Turkey, November 2019.
- [6] Oskar Maier, Bjoern Menze, Janina Gablentz, Levin Häni, Mattias Heinrich, Matthias Liebrand, Stefan Winzeck, Abdul Basit, Paul Bentley, Liang Chen, Daan Christiaens, Francis Dutil, Karl Egger, Chaolu Feng, Ben Glocker, Michael Götz, Tom Haeck, Hanna-Leena Halme, Mohammad Havaei, and Mauricio Reyes. Isles 2015 – a public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical Image Analysis, 35, 07 2016.
- [7] Sıĺvia Delgado Olabarriaga, Jean-Michel Rouet, Maxim Fradkin, Marcel Breeuwer, and Wiro J. Niessen. Segmentation of thrombus in abdominal aortic aneurysms from cta with nonparametric statistical grey level appearance modeling. IEEE Transactions on Medical Imaging, 24:477–485, 2005.
- [8] Parmar, P. Grossmann, J. Bussink, P. Lambin, and H. Aerts. Machine learning methods for quantitative radiomic biomarkers. Scientific reports, 5(13087), 2015.
- [9] Branch Coslett Peter E. Turkeltaub Nicholas Tustison Myrna F. Schwartz Pustina, Dorian and Brian Avants. Automated segmentation of chronic stroke lesions using linda : Lesion identification with neighborhood data analysis. Human Brain Mapping, 37(4):1405–1421, 2016.
- [10] Emmad Qazi, Alexis T Wilson, Connor McDougall, Mari Boesen, Fahad S Al-Ajlan, Pooneh Pordeli, Connor Batchelor, Khayam Khan, Tolulope T Sajobi, Ting-Yim Lee, Michael D Hill, Andrew M Demchuk, Mayank Goyal, Christopher D d’Esterre, Bijoy K Menon, and Nils D Forkert. Abstract tp45: One threshold does not fit all: Hounsfield unit thresholds to segment clot on ncct are patient specific. Stroke, 47(suppl 1):ATP45–ATP45, 2016.
- [11] Christian H. Riedel, Ulf Jensen, Axel Rohr, Marc Tietke, Karsten Alfke, Stephan Ulmer, and Olav Jansen. Assessment of thrombus in acute middle cerebral artery occlusion using thin-slice nonenhanced computed tomography reconstructions. Stroke, 41(8):1659–1664, 2010.
- [12] Emilie M. M. Santos, Henk A. Marquering, Olvert A. Berkhemer, Wim H. van Zwam, Aad vander Lugt, Charles B. Majoie, Wiro J. Niessen, and on behalf of the MR CLEAN investigators. Development and validation of intracranial thrombus segmentation on ct angiography in patients with acute ischemic stroke. PLOS ONE, 9(7):1–8, 07 2014.
- Date de l’appel : 13/12/2021
- Statut de l’appel : Pourvu
- Contacts cotés IBISC : Jean-Philippe CONGE (doctorant IBISC, équipe SIAM), Vincent VIGNERON (MCF HDR Univ. Évry, IBISC équipe SIAM), Hichem MAAREF (PR IUT Évry, IBISC équipe SIAM) {vincent.vigneron,hichem.maaref}@ibisc.univ-evry.fr, }
- Sujet de stage niveau Master 2 (format PDF)
- Web équipe SIAM